News
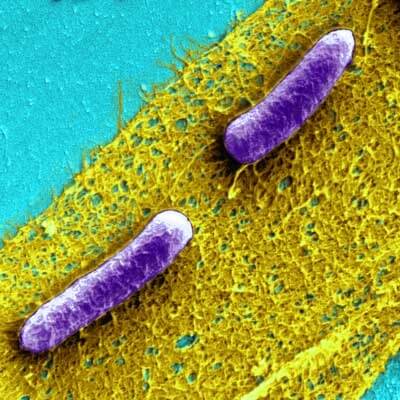
Biofilms are communities of bacteria (E. coli are depicted here in purple) ensconced in a slimy, but extremely tough matrix of extracellular material.
Cambridge/Boston, Mass. – September 17, 2014 – For most people, biofilms conjure up images of dirty drains and slippery stones in a stream bed. While there are plenty of “bad” biofilms around—they even cause pesky dental plaque and a host of other more serious medical problems—a team at the Wyss Institute for Biologically Inspired Engineering at Harvard University sees biofilms as a robust new platform for designer nanomaterials that could clean up polluted rivers, manufacture pharmaceutical products, fabricate new textiles, and more.
In short, they want to give biofilms a facelift, and have developed a novel protein engineering system called BIND to do so. Using BIND, which stands for Biofilm-Integrated Nanofiber Display, the team believes biofilms could be tomorrow’s living foundries for the large-scale production of biomaterials that can be programmed to provide functions not possible with existing materials. They have reported the proof of concept in Nature Communications.
“Most biofilm-related research today focuses on how to get rid of biofilms, but we demonstrate here that we can engineer these super-tough natural materials to perform specific functions—so we may want them around in specific quantities and for specific applications,” said the study’s senior author, Neel Joshi, who is an Associate Professor of Chemical and Biological Engineering at the Harvard School of Engineering and Applied Sciences (SEAS) and a Core Faculty member at the Wyss Institute.
Biofilms also self-assemble and self-heal. “If they get damaged, they grow right back because they are living tissues,” said lead author Peter Nguyen, a postdoctoral fellow at Harvard SEAS and the Wyss Institute.
Biofilms are communities of bacteria ensconced in a slimy, but extremely tough, matrix of extracellular material composed of sugars, proteins, genetic material, and more. During biofilm formation, individual bacteria pump out proteins that self-assemble outside the cell—creating tangled networks of fibers that essentially glue the cells together into communities that keep the bacteria safer than they would be on their own.
Interest in biofilm engineering is skyrocketing, and while several other teams have recently developed genetic tools to control biofilm formation, Joshi’s team has altered the composition of the extracellular material itself—essentially turning it into a self-replicating production platform to churn out whatever material they wish to produce.
“Until recently there was not enough cooperation between synthetic biologists and biomaterials researchers to exploit the synthetic potential of biofilms this way. We are trying to bridge that gap,” Joshi said.
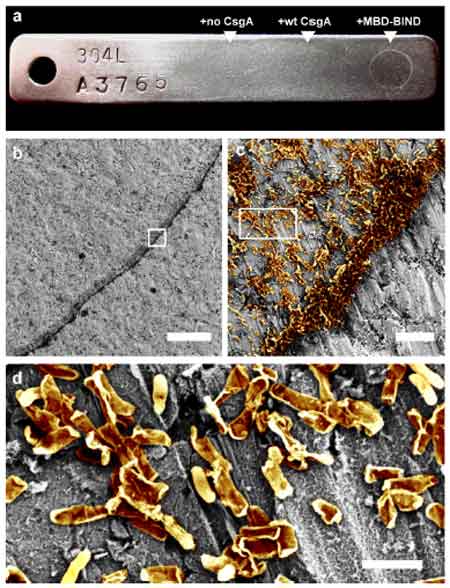 Joshi’s team has demonstrated the ability to program E. coli biofilms to stick to certain substrates, such as stainless steel. They fused a protein called MBD to the CsgA protein to develop a biofilm, which they allowed to adhere to steel over 48 hours—and then confirmed using a scanning electron microscope that the biofilm withstood a vigorous washing procedure (as shown in c-d).
Joshi’s team has demonstrated the ability to program E. coli biofilms to stick to certain substrates, such as stainless steel. They fused a protein called MBD to the CsgA protein to develop a biofilm, which they allowed to adhere to steel over 48 hours—and then confirmed using a scanning electron microscope that the biofilm withstood a vigorous washing procedure (as shown in c-d).
The team genetically fused a protein with a particular desired function—for example, one known to adhere to steel—onto a small protein called CsgA that is already produced by E. coli bacteria. The appended domain then went along for the ride through the natural process by which CsgA gets secreted outside the cell, where it self-assembled into supertough proteins called amyloid nanofibers. These amyloid proteins retained the functionality of the added protein—ensuring in this case that the biofilm adhered to steel.
Amyloid proteins traditionally get a bad rap for their role in causing tremendous health challenges such as Alzheimer’s disease, but in this case their role is fundamental to making BIND so robust. These amyloids can spontaneously assemble into fibers that, by weight, are stronger than steel and stiffer than silk.
“We are excited about the versatility of the method, too,” Joshi said. The team demonstrated an ability to fuse 12 different proteins to the CsgA protein, with widely varying sequences and lengths. This means in principle that they can use this technology to display virtually any protein sequence—a significant feature because proteins perform an array of impressive functions, such as binding to foreign particles, carrying out chemical reactions, transmitting signals, providing structural support, and transporting or storing certain molecules.
Not only can these functions be programmed into the biofilm one at a time, but they can be combined to create multifunctional biofilms as well.
The concept of the microbial factory is not a new one, but for the first time it is being applied to materials, as opposed to soluble molecules like drugs or fuels. “We are essentially programming the cells to be fabrication plants,” Joshi said. “They don’t just produce a raw material as a building block; they orchestrate the assembly of those blocks into higher-order structures and maintain that structure over time.”
“The foundational work Neel and his team are doing with biofilms offers a glimpse into a much more environmentally sustainable future where gargantuan factories are reduced to the size of a cell that we can program to manufacture new materials that meet our everyday needs—from textiles to energy and environmental clean-up,” said Don Ingber, founding director of the Wyss Institute, Judah Folkman Professor of Vascular Biology at Harvard Medical School and Children’s Hospital Boston, and Professor of Bioengineering at Harvard SEAS.
For now, the team has demonstrated the ability to program E. coli biofilms that stick to certain substrates, such as steel, and others that can immobilize an array of proteins or promote the templating of silver for construction of nanowires.
This work was funded primarily by the Wyss Institute for Biologically Inspired Engineering at Harvard University. The authors also acknowledge support from the National Science Foundation Graduate Research Fellowship and the A*STAR National Science Graduate Fellowship.
Topics: Bioengineering
Cutting-edge science delivered direct to your inbox.
Join the Harvard SEAS mailing list.
Press Contact
Caroline Perry